The aim of the National Resource for Network Biology (NRNB) is to advance the new science of Biological Networks through analytic tools, visualizations, databases and computing resources. Biomedical research is increasingly dependent on knowledge of biological networks of multiple types and scales, including gene, protein and drug interactions, cell-cell and cell-host communication, and vast social networks. Our technologies enable researchers to assemble and analyze these networks and to use them to better understand biological systems and, in particular, how they fail in disease. NRNB leverages the Cytoscape Cyberinfrastructure, a widely-used network analysis platform which we further support through a rich set of tutorials, workshops, 24/7 help desk assistance, and over 100 collaboration and service projects.
We are developing freely available, open source software technologies to enable network-based visualization, analysis, and biomedical discovery. We are driving research and development projects, coordinating training opportunities, and actively seeking new collaborations to develop NRNB tools and apply them to biomedical research.
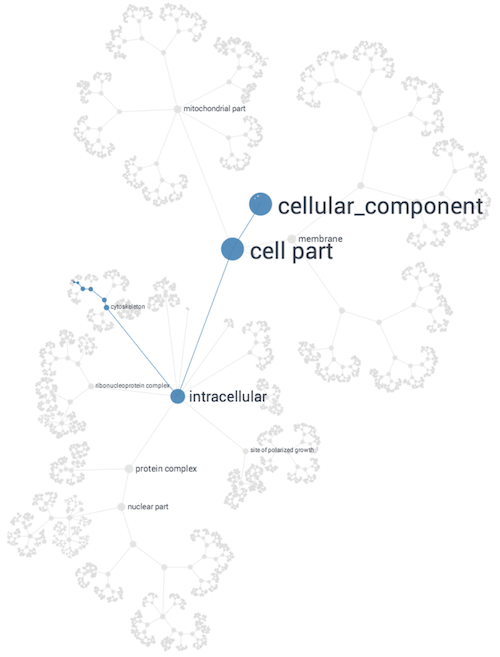
Network Approach to Building Gene Ontologies
NeXO (The Network Extracted Ontology) uses a principled
computational approach which integrates evidence from hundreds of thousands of individual gene and
protein interactions to construct a complete hierarchy of cellular components and processes. This
data-derived ontology aligns with known biological machinery in the GO Database and also uncovers
many new structures.

Network-based stratification of tumor mutations
NBS is a method for stratification (clustering) of patients in a cancer cohort based on somatic mutations and a gene interaction network. The method uses network propagation to integrate genome wide somatic mutation profiles for each patient over a gene interaction network, and a non-negative matrix factorization based clustering approach in order to derive biologically meaningful stratification of a patient cohort.
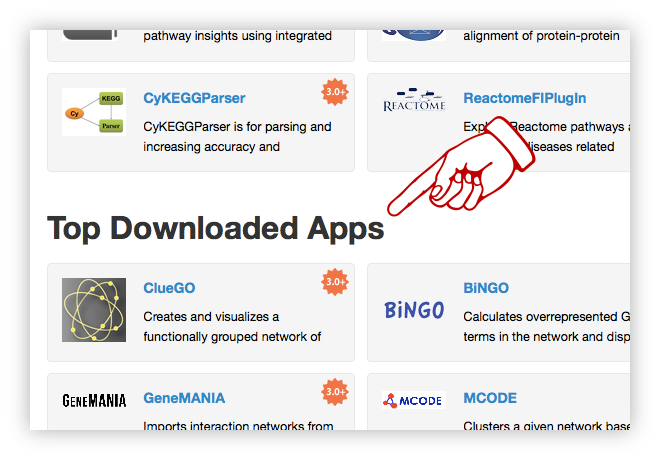
Cytoscape App Store
The overarching goals of the Cytoscape App Store are to
highlight the important features apps add to Cytoscape, to enable researchers to find apps they
need, and for developers to promote their apps. The Cytoscape App Store highlights the important
features that apps add to Cytoscape, enable researchers to find and install apps they need, and help
developers promote their apps.
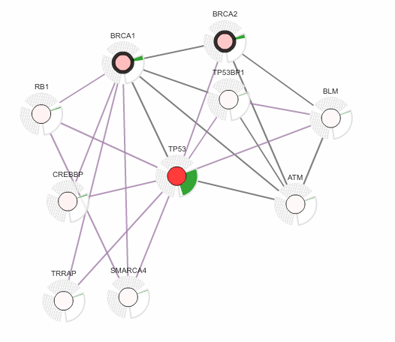
Network Analysis Tools
Our research programs are exploring an array of new approaches to network analysis, such as how to
use
molecular networks to stratify cancer patient genomes (Network-Based Stratification) or how to
assemble
a gene ontology of the cell directly from molecular network data (nexontology.org). In recent years
we
have released a number of widely-used network analysis resources, including the Cancer Biology
Portal
(cBioPortal.org), the GeneMANIA network
database (genemania.org), and the Cytoscape Appstore
(apps.cytoscape.org).